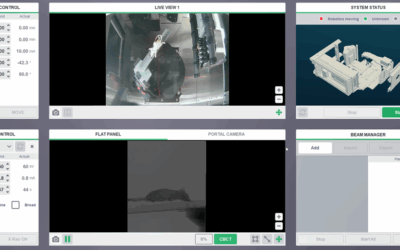
Small animal x-ray irradiation platforms are expanding the capabilities and future pathways for radiobiology research. Meanwhile, proton radiotherapy is transitioning to a standard treatment modality in the clinician’s precision radiotherapy toolbox, highlighting a gap between state-of-the-art clinical radiotherapy and small animal radiobiology research. Comparative research of the biological differences between proton and x-ray beams could benefit from an integrated small animal irradiation system for in vivo experiments and corresponding quality assurance (QA) protocols to ensure rigor and reproducibility. The objective of this study is to incorporate a proton beam into a small animal radiotherapy platform while implementing QA modelled after clinical protocols. A 225 kV x-ray small animal radiation research platform (SARRP) was installed on rails to align with a modified proton experimental beamline from a 230 MeV cyclotron-based clinical system. Collimated spread out Bragg peaks (SOBP) were produced with beam parameters compatible with small animal irradiation. Proton beam characteristics were measured and alignment reproducibility with the x-ray system isocenter was evaluated. A QA protocol was designed to ensure consistent proton beam quality and alignment. As a preliminary study, cellular damage via γ-H2AX immunofluorescence staining in an irradiated mouse tumor model was used to verify the beam range in vivo. The beam line was commissioned to deliver Bragg peaks with range 4-30 mm in water at 2 Gy min-1. SOBPs were delivered with width up to 25 mm. Proton beam alignment with the x-ray system agreed within 0.5 mm. A QA phantom was created to ensure reproducible alignment of the platform and verify beam delivery. γ-H2AX staining verified expected proton range in vivo. An image-guided small animal proton/x-ray research system was developed to enable in vivo investigations of radiobiological effects of proton beams, comparative studies between proton and x-ray beams, and investigations into novel proton treatment methods.
Kim MM, Irmen P, Shoniyozov K, Verginadis II, Cengel KA, Koumenis C, Metz JM, Dong L, Diffenderfer ES.